
ERVs and common descent

ERVs and common descent
![]()
A significant problem in
a
nutshell with the orthologue
argument for ERVs as evidence of common descent
(taken from here)
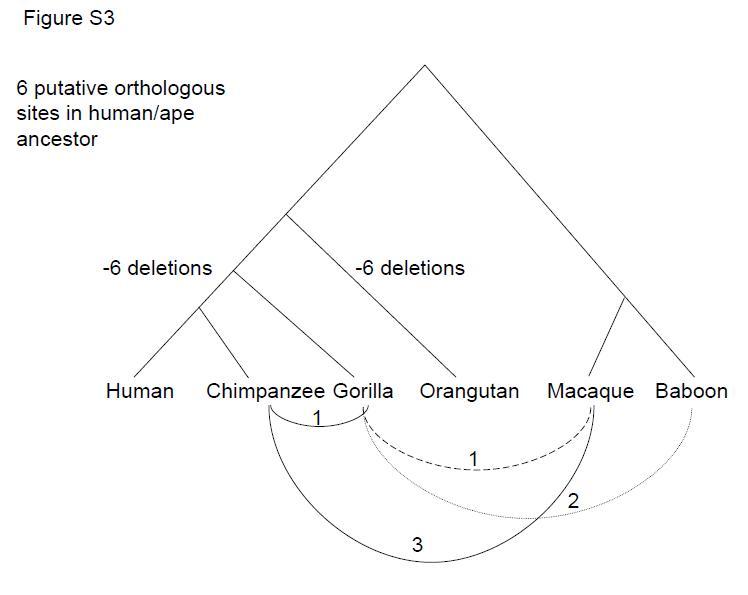
(Incidentally, as the author
acknowledges, the no. of missing ERVs in man on the left should be 7,
of course,
not 6 as is correct for the Orangutan.) The same problem
of missing deletions from potentially orthologous sites also applies to
CERV 2. Despite some
intensive searching, I've only been
able to
find two candidate orthologous CERV 1 sites one Macaque to Pan Trog.
(NW_001112574.1 vs. Contig33.125) and one Gorilla to Pan. trog.
(AADA01328632.1 vs CABD02058267.1), and
neither is anything like as persuasive as SA Smith's example for CERV 5
(post 193) in her
blog taken from here.
It has opened my eyes to the complexity and difficulty of the
search though. Several
interesting findings including genome-wide distribution of a
non
CERV fragment of NW_001112574.1 near to a CERV 1 site
(coords 7346538-7350059) in Pan. trog., Homo sap., Macaque malatta (the
origin of the sequence), Orangutan and Gor. gorilla. Image of blast
hit results:
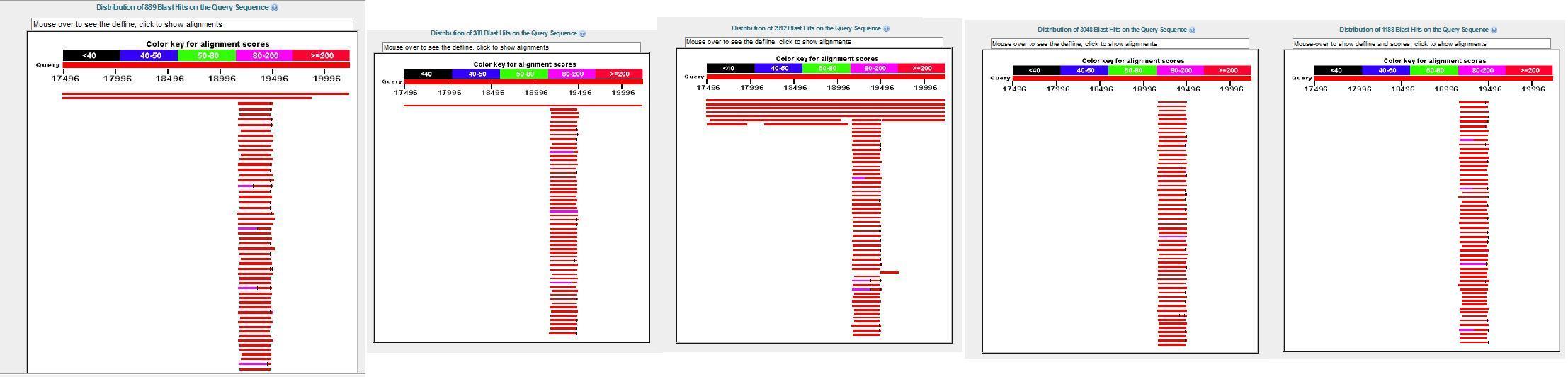
A few details:
I searched for the pol gene of CERV1 (4544..5611 from NCBI's
AY692036.1) in the macaque genome and then took contiguous sequences of
the hits from this with adjacent non CERV containing sequence and
blasted it against Pan Trog.
In the macaque contig NW_001112574.1 the a portion of CERV1 pol gene is
found at 7352782-7351802(-), (though there's a gap in pol sequence from
its 487th to 664th nucleotide).
Two non CERV containing adjacent fragments (I called a and b) from the
macaque contig are also with the pol gene in the chimp contig for
chromosome 2, Contig33.125 (AACZ03012828.1).
The fragments a and b are found at 7347266-7347577 and 7349461-7349794
in NW_001112574.1. None of 94 other chimp contigs shared either of
these two non-CERV fragments from the macaque genome and CERV1 pol
sequence.
A third non CERV fragment from the same macaque contig (which I called
c) 7346538-7350059, near the pol sequence, was not in the chimp contig,
but showed widespread distribution throughout the genome of several
primates (nearly 3 k hits in man and 3,048 in orangutan). It was inside
the regions of 15 different macaque genes.
The second interesting site was shared between the gorilla contig
CABD02426596.1 and chimp AADA01328632.1 . Most of the shared sequence
is from CERV1, but there is a non CERV1 region from 6129 to 6395 in the
gorilla sequence also found in the chimp sequence at 800 to 1065 (Blast
scores: 375 375 100% 3e-107 92%).
Frustrating
and fascinating as this is, I lack the time and expertise to take the
matter
further presently.
Some
other interesting recent reflections on ERVs here
and here.
